Czii Object Identification
Overview
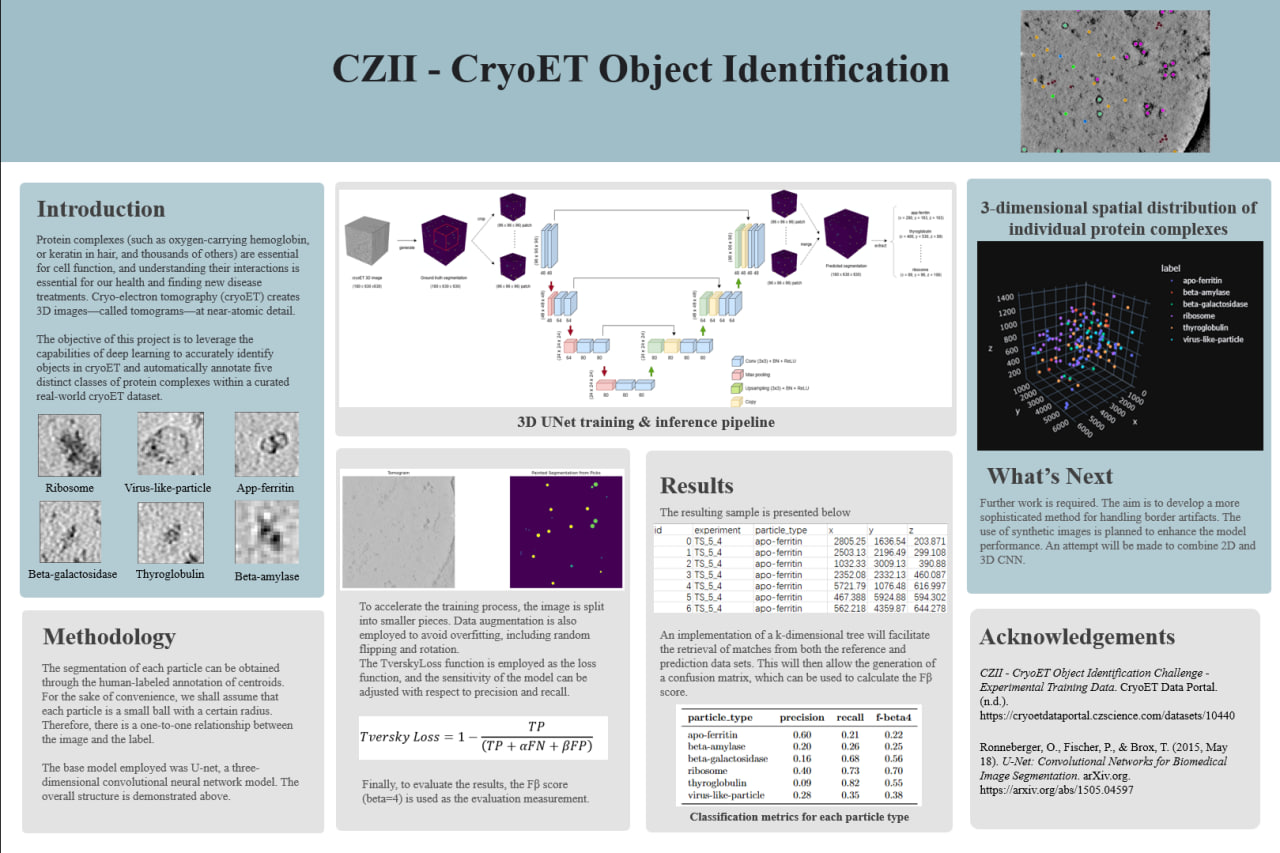
This project applies deep learning techniques to Cryo-Electron Tomography (CryoET) data, enabling the precise identification and segmentation of protein complexes. It automates the annotation process, addressing the challenges of low signal-to-noise ratios and dense cellular environments, advancing biological research and medical discoveries.
Dataset
The dataset includes seven CryoET tomograms (180×630×630) with manually annotated centroids for six protein complexes:
- Protein Types: Apo-ferritin, beta-amylase, beta-galactosidase, ribosomes, thyroglobulin, and virus-like particles.
- Challenges: High data complexity and noise requiring advanced preprocessing and augmentation.
Methodology
- Deep Learning Models: Implemented a 3D U-Net architecture for voxel-level segmentation, with Tversky loss to balance precision and recall.
- Patch-Based Training: Divided tomograms into 96×96×96 patches to reduce computational load and optimize training efficiency.
- Centroid Localization: Utilized connected component analysis and KD-trees for accurate object identification within reconstructed tomograms.
Key Findings
- Precision: Achieved high precision for ribosomes and apo-ferritin (up to 0.72).
- Recall: Maintained near-perfect recall across most protein types, ensuring minimal missed detections.
- Metrics: fbeta-4 score peaked at 0.72, balancing recall and precision for dense particle localization.